Monoqlo℠
◄ Back to Open Source @ NYSCFBackground
The New York Stem Cell Foundation Research Institute (NYSCF) has built a deep learning framework for automatically detecting clonalized cell colonies and assessing clonality from daily imaging data. Our approach, Monoqlo℠, features multiple CNN “modules” for both detection and classification, which are integrated and automatically deployed using Python scripts.
Full details on the design and rationale of the Monoqlo℠ framework are provided in the article linked here.
NYSCF GitHub Repository
The NYSCF GitHub repository contains all of the code necessary for executing the Monoqlo℠ framework using an example dataset.
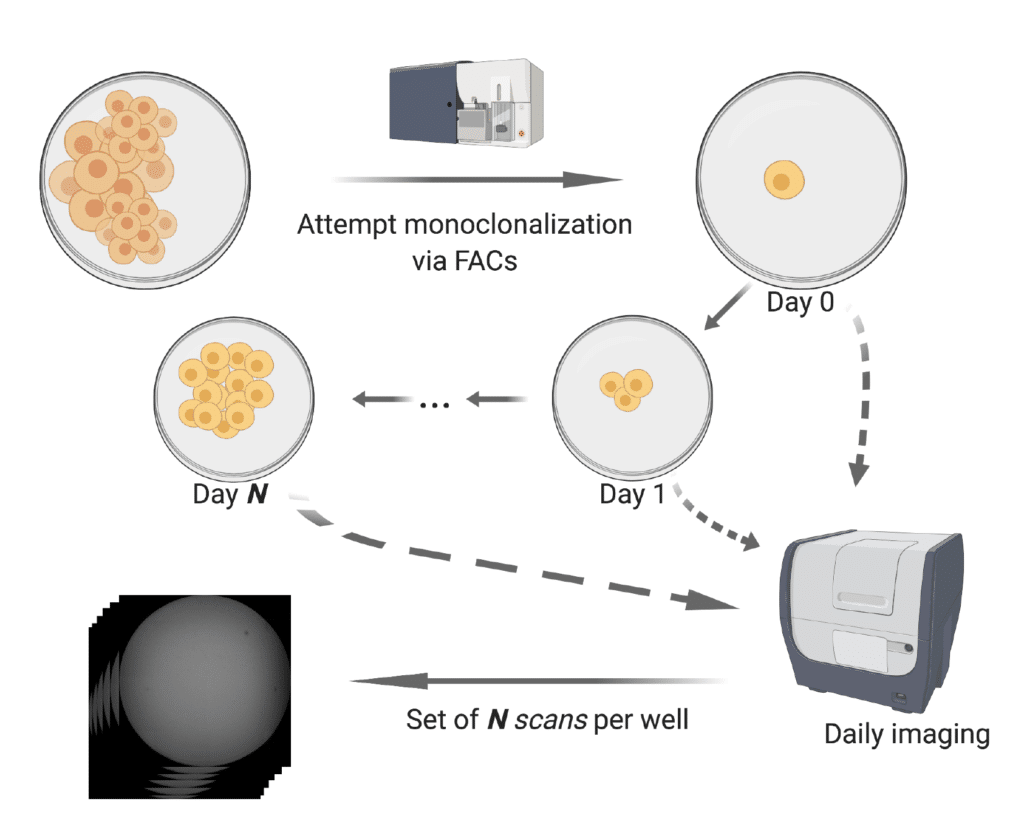
Monoqlo℠ Imaging Dataset
Below we provide a demonstrative example of an imaging dataset, which includes an illustration of the Monoqlo℠ framework’s execution logic and performance.
DMR0001 represents the image set for an entire, real-world monoclonalization run which has been fully deidentified, consisting of daily scans for each well of 8 96-well plates.
Note: this dataset is >160 GB in size
Monoqlo Dataset (DMR0001) by NYSCF is licensed under CC BY-NC-SA 4.0

Disclaimer
The code and example datasets provided are intended as a demonstration of the Monoqlo℠ framework’s capabilities and general uses only, without any representations or warranties. Depending on imaging modality and requirements for use, it may be necessary to train new models and/or adapt the source code for execution on your own data.